To verify the feasibility of the project, experiments were designed for each of the four modules to verify the proper functioning of the gene circuits within Chlamydomonas reinhardtii.
Infection Report
Module
1. Plasmid Construction
Use Golden Gate assembly:
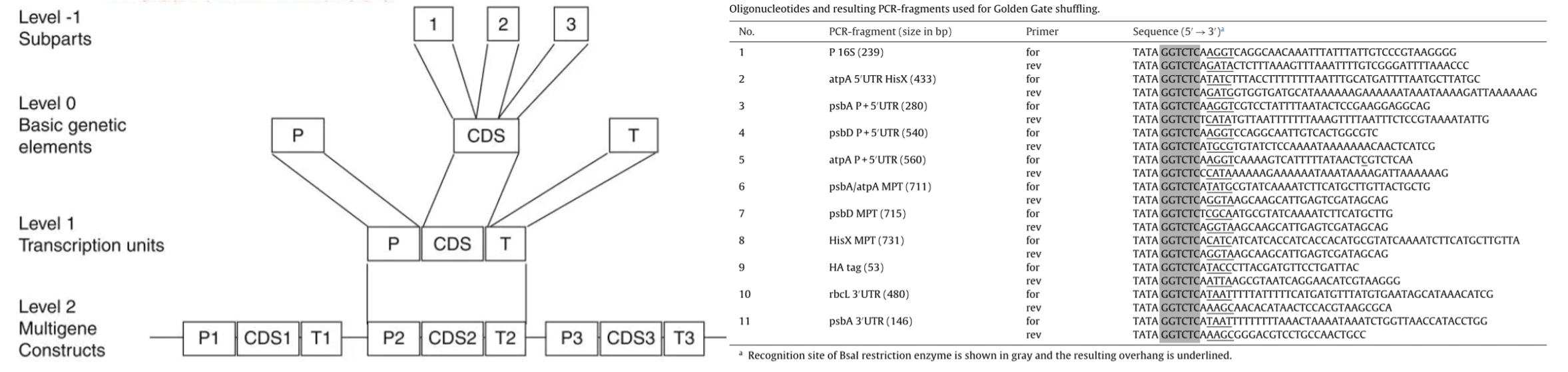
Level_-1:
Introduce the respective primers and BsaI recognition sequences (GGTCTCN,which was avoided in the design of Parts) to the 5' end of the target fragment ,including crLasR-HA_Tag, aadA, mRFP1, mbal-HA_Tag, aphA6, respectively by PCR in ligation order to form the assembly subunit.

Level_ 0:
Assemble the structural genes into CDS in sequence, prepare the promoter (including 5'UTR) ,RBS and terminator, awaiting for further assembly into circuits.
The fusion sequence uses the Level_0 prefix and suffix as specified in the iGEM RFC 1000 standard below[2].
| Fusion Site 5' | Part Type | Fusion Site 3' |
|---|---|---|
| GGAG | Promoter | TACT |
| TACT | 5'UTR | AATG |
| AATG | CDS | GCTT |
| GCTT | Terminator | CGCT |
Level_ 1:
Complete the assembly of the basic circuit, including the two gene circuits mentioned in the Design section and one circuit for testing the function of chromoproteins;
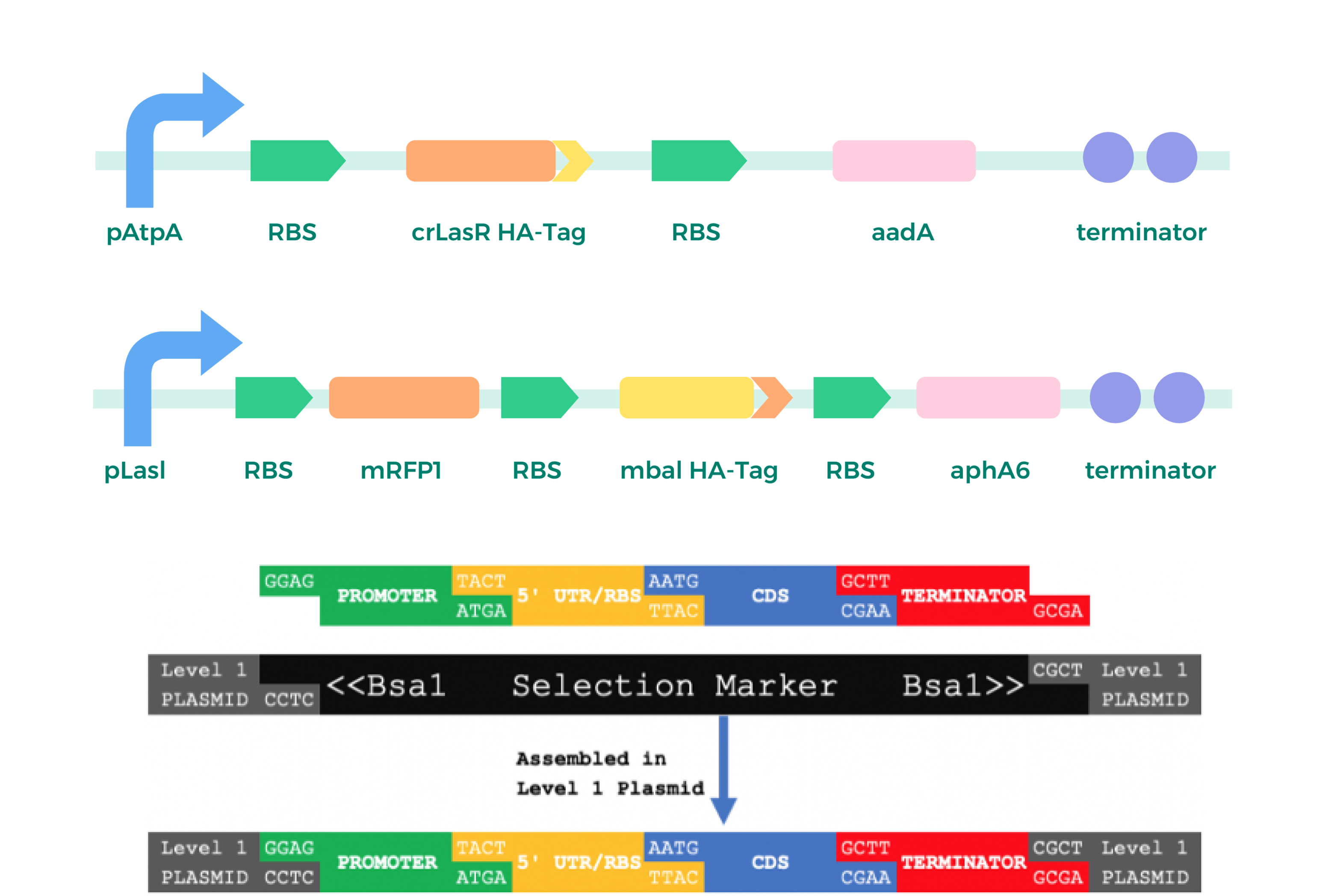
Level_ 2:
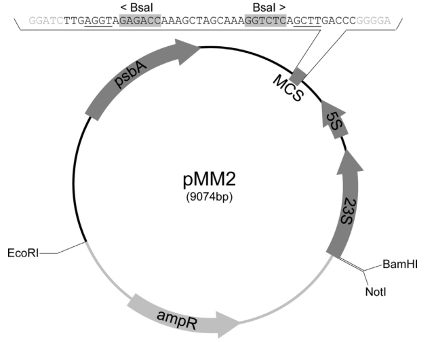
Each of the three circuits are loaded into MCS in the pMM2 chloroplast vector (using the BsaI site as well)[1] to form a mature plasmid ready for the next step of transformation.
2. Chloroplast Transformation and Testing
in Chlamydomonas Reinhardti
Fud7 recipient strains were obtained from the Chlamydomonas Resource Center, cultured in TAP medium (Table 1) until the late exponential growth phase, and concentrated cells were collected using polyamide filters. Then chloroplast transformation was performed with tungsten particles loaded with transformation plasmids [1](gene gun method, as shown in the figure below).
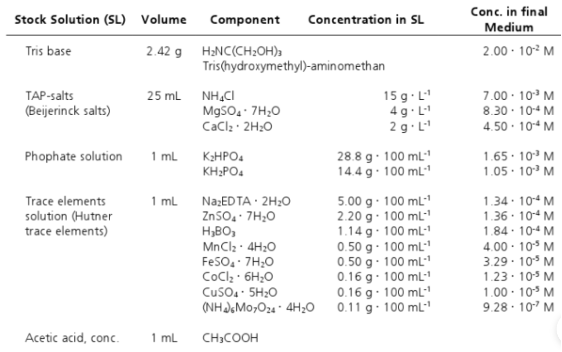

Upon transformation of the plasmid into chloroplasts, our genes of interest (GOI) will recombine with homologous segments of the chloroplast genome due to the psbA,5S,23S gene homologous sequences carried in the pMM2 sequence itself, resulting in a stable transformation.[1]
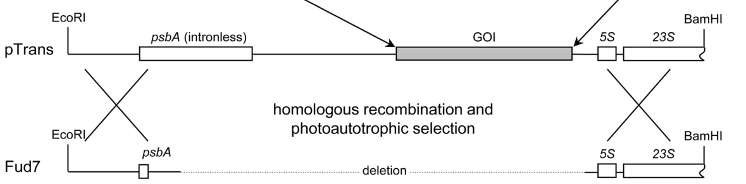
Testing method:
- (i) Testing for successful entry of the plasmid into the cells based on the ampR (ampicillin resistance gene )carried on the pMM2 plasmid.
- (ii) Testing for successful loading of GOI based on the spectinomycin and kanamycin resistance.
- (iii) Testing for successful protein expression based on HA-Tag immunoprecipitation.
The Chlamydomonas chassis obtained by all the above screening were transferred from TAP medium to HS liquid medium and cultured under continuous light for several generations until the desired homogeneity was formed[1].
3. Device and Genetic Circuits Function Testing
a) CrLasR Protein Expression Assay
Without AHL stimulation, total proteins of Chlamydomonas cells were extracted at different growth times, and then identified crLasR protein expression using Anti-HA magnetic beads and plotted “protein-time relationship curves”.(as shown below)
b) CrLasR and AHL Interaction Assay
At the peak time of crLasR expression in a), set different AHL concentration gradients for induction, and pLasI-GFP circuits in easy-to-operate Saccharomyces cerevisiae was constructed to plot “fluorescence intensity-AHL concentration curves”.
c) Chromogranin Function Assay
(i) Test 1: Expression Assay
Following the introduction of the plasmid loaded with PAtpA-RBS-mRFP1-terminal, antibiotic screening was performed to observe whether the colonies were red for checking if mRFP1 was properly expressed in Chlamydomonas chloroplasts.
(ii) Test 2: Response Assay
Measure and plot the “chromogranin signal intensity-time curves” in Chlamydomonas reinhardtii, under the condition of the peak time for crLasR expression in a) and the concentration of AHL to lead maximum fluorescence intensity in b).
(iii) Test 3: Effect Assay
All else being the same, a suitable perceptible signal intensity of the curve in (ii) was selected as the threshold, and then replace the type of chromoprotein in the circuit and experimentally determine the time taken for different chromoproteins to reach the threshold for report optimization.
Quorum Sensing(QS)
Module
1. Vector Construction and Transformation
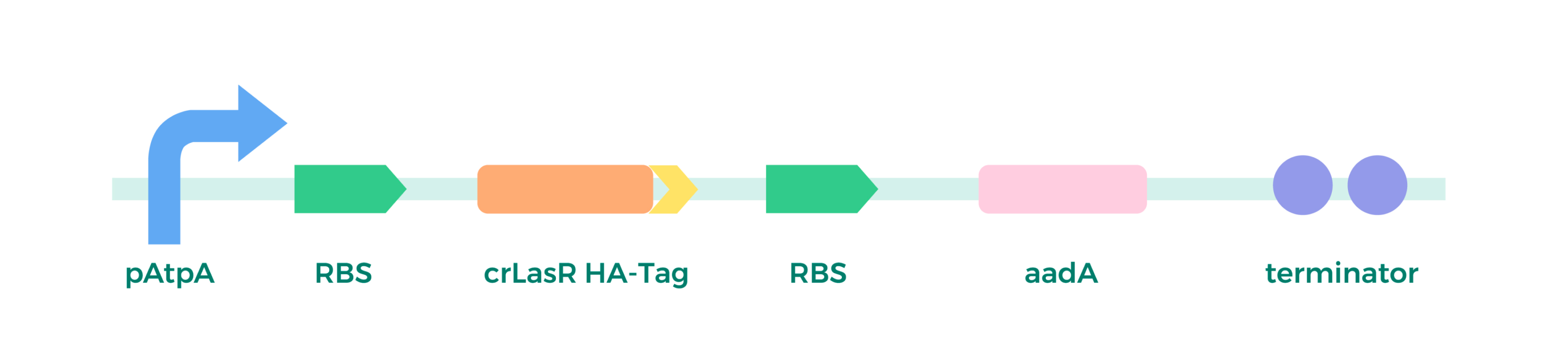
a) Plasmid Construction:
relying on Golden Gate/Mocol cloning, assembly method as above.
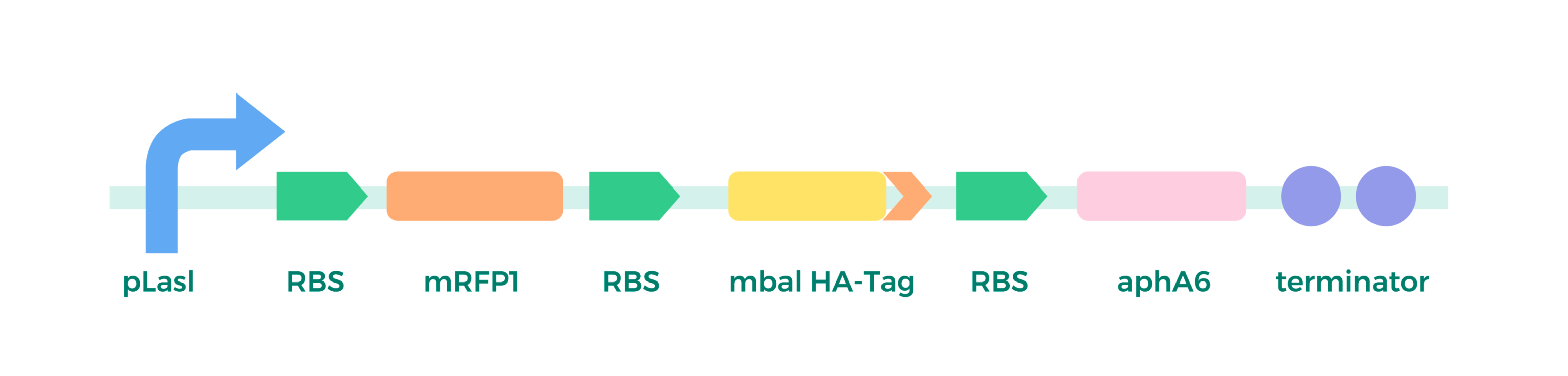
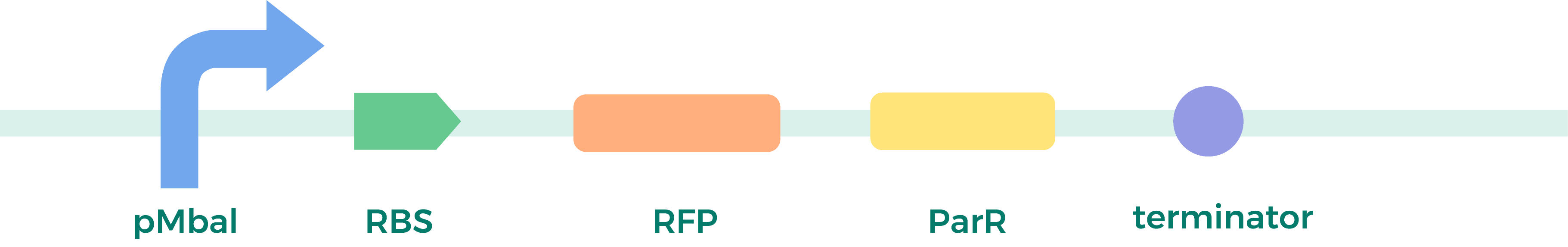
b) Chloroplast Transformation and Detection
in Chlamydomonas Reinhardtii
i. The transformation method as above, omitted.
ii. Detection:
- 1. Test for successful entry of plasmids into cells based on ampR reporter genes carried on pMM2 plasmids.
- 2. The MbaI_express pathway was successfully transferred in with kanamycin and the pLasI promoter was functioning normally; the MbaR_express pathway was successfully transferred in with thaumycin and the PsaD-tVP6 promoter was functioning normally; the pMbaI_promote pathway was successfully transferred in with balomycin and the pMbaI-cycmini100 promoter was functioning normally.
- 3. Extract total proteins from Cladosporium reinhardtii that have been successfully transferred into plasmids and have a functioning pathway.
iii. Verify the expression of MbaI and MbaR by Westen blot (with antibodies to His-tag and HA-tag, respectively) and the expression amount. Protein expression was screened for promoters, terminators and other components.[8]
c) Plasmid Amplification
2. Component and Circuit Function Testing
a) Validation of the population-sensing pathway in response to AHL2 (3-OH-C10-HSL).
Experiments were performed in Chlamydomonas reinhardtii chassis transferred to lines MbaR_express and pMbaI_promote, and engineered Chlamydomonas reinhardtii was cultured in liquid medium. Set up different gradients of AHL2 molecule concentrations for induction, measure the fluorescence intensity in an enzyme marker and make an AHL2 molecule concentration-fluorescence intensity curve. The MbaR_express and pMbaI_promot lines can be verified in the easy-to-operate Saccharomyces cerevisiae first.
b) Validation of the population-sensing pathway in response to AHL1 (3-oxo-C12-HSL).
Experiments were performed in Chlamydomonas reinhardtii sump transferred to lines LasR_express, MbaI_express, MbaR_express and pMbaI_promote, and engineered Chlamydomonas reinhardtii was cultured in liquid medium. Different gradients of AHL1 molecule concentration were set for induction, and the fluorescence intensity was measured in an enzyme marker to make an AHL1 molecule concentration-fluorescence intensity curve. The resulting curves are compared with the response curves of the Chlamydomonas reinhardtii sump to the AHL1 concentration by transferring to the bacterial sensing pathway without coupling to the population sensing pathway to assess the amplification effect of the population sensing pathway. The same can be verified in yeast first.
c) Validating the effect of a population sensing system to unify the expression of the Rhizobium population.
Experiment in a Chlamydomonas reinhardtii sump transferred to the lines LasR_express, MbaI_express, MbaR_express and pMbaI_promote by culturing engineered Chlamydomonas reinhardtii on solid medium. A concentration gradient of AHL1 molecule solution was added at one end of the medium and the time until the whole medium yeast fluoresced was observed and recorded for the AHL1 molecule concentration-time curve. The same can be verified in yeast first.
Note
Pseudomonas aeruginosa 3-oxo-C12-HSL, Shanghai Yuanye Biotechnology, item. S43478; M. tundripaludum 21/22 3-OH-C10-HSL, Kemegen (Shanghai) Biotechnology, item. CY16482.
d) Verification that the AHL2 molecule produced by Chlamydomonas reinhardtii does not affect the stability of the system
i. Verify whether AHL2 molecules affect the behavior and morphology of Chlamydomonas reinhardtii
Experiments were conducted in Chlamydomonas reinhardtii without engineering modifications. An appropriate amount of AHL2 molecules was added to the experimental group and an equal amount of buffer solution was added to the control group, and observations were made for both groups of Chlamydomonas reinhardtii.
1. Chlamydomonas growth measurement[6]
The algae were collected under aseptic conditions using the turbidity method, and the OD750 values of the algal solution were measured using a UV-Vis spectrophotometer (Pu-Analysis, Beijing).
2. Determination of cytochrome content of Chlamydomonas reinhardtii[5,6]
The contents of chlorophylls a and b and carotenoids of Chlamydomonas reinhardtii were determined by spectrophotometry. After removing the supernatant, the algal cells were extracted twice with 90% acetone in the dark for 24 h. The extracts were finally fixed to 10 m L. The OD values of the extracts were measured at 665, 649 and 470n using a UV-Vis spectrophotometer. The chlorophyll a content (Chla), chlorophyll b content (Chlb), and carotenoid content were calculated as follows (Sartory and Grobbelaar 1984):
3. Determination of chlorophyll fluorescence parameters[5]
The photochemical activity of algal cells was measured using a multi-excitation wavelength modulated chlorophyll fluorometer (Walz, Germany). The fluorescence parameters were measured by sampling each group every day. The assay procedure was as follows: first, the culture of Chlamydomonas reinhardtii at the exponential growth stage was darkened for 12 h. Then, 2 m L of the algal solution was added to a quartz cup, the measurement light (ML) wavelength was set to 440 nm, the current fluorescence level Ft was adjusted to 1.5, the script sigma 500 chl and performed O-I1 fitting, saved the optical cross-sectional area data of Chlamydomonas reinhardtii, performed fast light curve in sp-analysis mode, applied the saved optical cross-sectional area data to the fast light curve, and finally recorded the initial (F0) and maximum (Fm) values of chlorophyll fluorescence, the actual photometric yield (YII) and the relative electron transfer efficiency (r ETR) in view-mode mode. electron transfer efficiency (r ETR).
ii. Verification of whether AHL2 molecules affect the behavior and morphology of wound bacteria
The common wound bacterium Pseudomonas aeruginosa was selected for experimental investigation. In the experimental group, appropriate amount of AHL2 molecules were added to the bacterial suspension and equal amount of buffer solution was added to the control, and the two groups of bacteria were observed. For Gram-negative bacteria, AHL reaches a certain concentration to activate relevant transcriptional regulators, synthesize extracellular polysaccharides, virulence factors and alginates, etc., which cause bacteria to aggregate and form biofilms. The self-inducible molecule receptor Lux R can respond to exogenous AHL by upregulating the expression of the bme B efflux pump and developing resistance to antibiotics.[7]
iii. Verify whether AHL2 molecules cause airborne bacterial tropism leading to gel contamination
1. Pseudomonas aeruginosa biofilm formation rate
2. Tropism of Pseudomonas aeruginosa to AHL2
3. Pseudomonas aeruginosa drug resistance (including antibiotics and antimicrobial peptides)
Four groups of experiments were set up: gel A with AHL2 molecules and encapsulated antimicrobial peptide; gel B without AHL2 molecules and encapsulated antimicrobial peptide; gel C with AHL2 molecules and no encapsulated antimicrobial peptide; and gel D without AHL2 molecules and no encapsulated antimicrobial peptide. the contamination rate of the four groups of gels was counted to assess whether the expression of AHL2 molecules would affect the infection process of the gels.
Treatment
Module
Plasmid construction and transformation
same method as the previous module
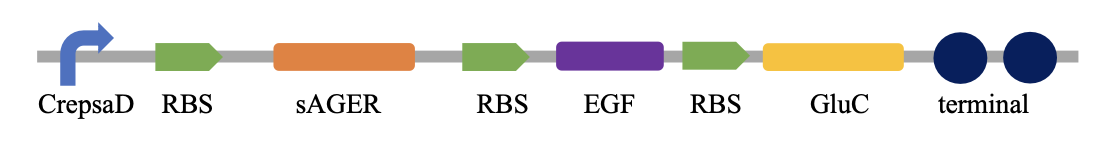
The inspection of component and line function
1. AGER protein secretion efficiency assay
a) In vitro preparation of AGE solution with known concentration and AGE receptor protein solution with known concentration, use ELISA double antibody sandwich method to produce a standard curve for the receptor protein.
A solid phase antibody was made by coating the microtiter plate with purified human glycosylation end product (AGE) antibody, adding glycosylation end product (AGE) sequentially to the microtiter wells coated with the monoclonal antibody, and then combining with HRP-labeled glycosylation end product (AGE) antibody to form an antibody-antigen-enzymatic antibody complex. TMB is converted to blue by HRP enzyme and to a final yellow color by the action of acid. The color shade is positively correlated with the glycosylation end product (AGE) in the sample. The absorbance (OD) was measured at 450 nm using an enzyme marker, and the concentration of human glycosylation end products (AGE) in the sample was calculated from the standard curve.
b) After the positive cell line with successful plasmid transduction was cultured in suspension, the absorbance value was taken from the suspension and the cell density was calculated; the supernatant was obtained by low-speed centrifugation of the suspension (only the proteins secreted to the extracellular were collected) and the efficiency of AGER protein secretion in the supernatant was determined.
2. Therapeutic effect of AGER protein on diabetic wounds
a) Take fresh blood from rats in diabetic model, clot naturally for 10-20 minutes at room temperature, and centrifuge for about 20 minutes (2000-3000 rpm).
b) The supernatant was carefully collected and a portion of the sample was taken to measure the serum AGE assay/serum protein content.
3. Human epidermal growth factor EGF protein expression efficiency
a) Select positive Cladosporium by PCR and inoculate them in 20mL of fresh TAP medium. The absorbance of 10mL of the medium was measured and the density of Chlamydomonas reinhardtii was estimated.
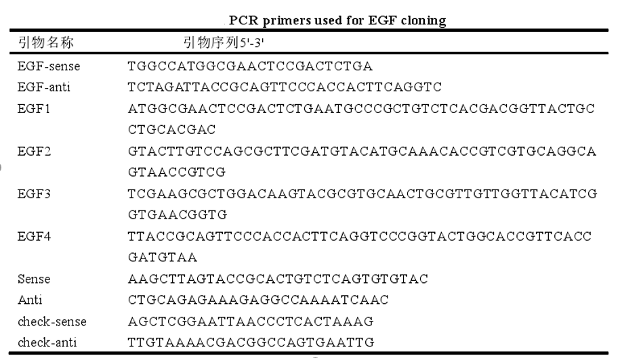
b) Centrifuge the supernatant at low speed (only the EGF protein secreted into the extracellular space is detected). Two groups of untransformed Chlamydomonas reinhardtii were used as negative controls.
c) Human EGF precoated ELISA kit was used to detect the content of EGF protein, and finally the OD450 was measured under spectrophotometer after the reaction between samples and reagents.
4. Measurement of luciferase expression efficiency
a) The positive Chlamydomonas cell line was cultured in liquid suspension, and the supernatant was taken after low-speed centrifugation to determine the amount of luciferase successfully secreted into the extracellular space.
b) The supernatant was mixed with D-luciferin standard solution in a specific ratio and the fluorescence intensity was analyzed by ImageJ software or fluorescence spectrophotometer.
c) The fluorescence intensity of the hydrogels successfully encapsulated with positive Cladosporium cells and luciferin was analyzed, and the optimum luciferin concentration was explored by changing the concentration of luciferin encapsulated in the hydrogels.


Biosafety
Module
Plasmid construction and transformation were performed as above.
1. Ferric Ion Excited Promoter Expression Assay
Experimental protocol: After linking the promoter ATX1 and the reporter gene RFP, the promoter was introduced into the plasmid to construct the expression vector for testing. Another group was introduced into the empty plasmid. Each set of experiments was repeated to verify the generalizability of the findings. The introduced Chlamydomonas reinhardtii was cultured in TAP medium, and AMP was added to the medium to screen the engineered strains for the introduced plasmids.
Steps:
- a) Divide Chlamydomonas reinhardtii into three groups, one control group and two experimental groups.
- b) The control group was introduced with a blank plasmid and the experimental group was introduced with a plasmid containing the ATX1-GFP sequence.
- c) Using Amp to screen out the successfully introduced Chlamydomonas reinhardtii
- d) The experimental groups were divided into two groups, one lacking Fe3+ and one containing Fe3+.
- e) The sensitivity of the promoter to Fe3+ was determined by observing the fluorescence status or the protein bands by SDS-PAGE gel electrophoresis.
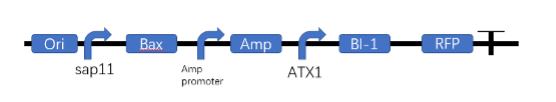
2. The ferric ion-excited promoter initiates the transcription of Bl-1 gene
The Bl-1 gene was conjugated with a ferric ion-initiated promoter, and the related sequence was introduced into the plasmid and then the expression vector was introduced into Chlamydomonas reinhardtii. The BL-1 protein was purified by isolation and then subjected to gel electrophoresis. The expression of the reaction protein was obtained by viewing the protein strips.
3. Test whether Bax toxin works properly in Chlamydomonas reinhardtii
Experimental protocol: ATX1-BAX sequence was constructed, inserted into the plasmid, and introduced into Chlamydomonas reinhardtii in TAP medium containing iron ions, and the control group was introduced into the blank plasmid. The successfully introduced strains were screened by Amp. The screened Chlamydomonas reinhardtii and the control were transplanted to TAP medium without iron ions, and the survival of the colonies was observed to determine whether the BAX toxin was working.
4. Detection of the working of gene lines
Experimental protocol: The experimental Chlamydomonas reinhardtii was divided into control and experimental groups, and the specific groups were set up as follows.
| Group | SAP11 | BAX | ATX1 | BL-1 | Fe3+/th> |
|---|---|---|---|---|---|
| NC_1 | - | - | - | - | + |
| NC_2 | - | - | - | - | - |
| Experimental 1 | + | + | - | - | - |
| Experimental 2 | + | + | - | - | + |
| Experimental 3 | - | - | + | + | - |
| Experimental 4 | - | - | + | + | + |
| Experimental 5 | + | + | + | + | - |
| Experimental 6 | + | + | + | + | + |
The group experiments were used to determine whether the individual components were functioning properly. The survival status of Chlamydomonas reinhardtii was observed to determine whether BL-1 in Chlamydomonas reinhardtii was able to inhibit the action of BAX toxin properly and act as a detoxification function. By isolating and purifying the protein, it was determined whether the iron-inducible promoter could work properly in the whole system. Finally, we will determine whether the iron-deficient state can kill Chlamydomonas reinhardtii properly and play a role in controlling biosafety.
Opretation Steps:
- 1. Assemble the plasmid using a biomodule conforming to RFC10 or Golden gate protocol
- 2. Introduce the plasmid into Chlamydomonas reinhardtii in TAP medium by electrotransformation
- 3. culture under suitable conditions
- 4. Observe and record the survival of Chlamydomonas reinhardtii cells
- 5. purify the proteins after 72H
- 6. SDS-PAGE of the isolated proteins and observation of the bands
Chassis Related
Methods
Chlamydomonas Growth Status Assessment
1. Determination of growth curve of Chlamydomonas reinhardtii
a. One ring of Chlamydomonas reinhardtii was inoculated in liquid medium and incubated in a light incubator.
b. The temperature is 20°C, the photoperiodic ratio is 16h/8h, the light intensity is 12,000Lx, and the speed of the shaker is 120r/min.
c. Samples were taken every 1 d. The blank control was a liquid medium without Chlamydomonas inoculum, and the OD values were measured at 395 nm wavelength.
d. Using the measured OD value as the vertical coordinate and the time interval of 1d as the horizontal coordinate, the growth curve of Chlamydomonas reinhardtii was plotted to reflect the growth of Chlamydomonas reinhardtii.
2. Measurement of optical density of Chlamydomonas cells
Two methods were used to determine the cell density values of Chlamydomonas reinhardtii.
a. Automatic monitoring using the Algal Station online monitoring system, with samples taken every 20 min
b. Offline verification with UV/Vis spectrophotometer at 750 nm, Jasco V-530, diluting the sample to stabilize OD750 below 1.0, then calibrating the OD7S0 value with the OD value detected by AS at the same time and converting the OD value detected by AS to OD750 using the calibration curve.

3. Specific Growth Rate $\mu$
$OD_2$ and $OD_1$ are the absorbance of AS at 12h intervals at the time of sampling, respectively.
4. Cell dry weight density determination
3-20 mL of cultured algal solution was centrifuged at 4000 rpm-5 min, the supernatant was discarded, washed with ultrapure water, filtered onto a pre-dried and weighed glass fiber membrane, dried at 60°C for 24h and then weighed. The formula was calculated as follows.
$w1$ represents the mass of the unfiltered membrane after drying ($g$), $w2$ represents the mass of the filtered membrane after drying ($g$), and v represents the volume of filtered algal fluid ($L$).
The yield of biomass at each stage of the incubation process was calculated as follows:
where $DW1$ and $DW2$ represent the dry weight ($g·L^{-1}$) for the corresponding interval of 12 h. $P$ is the yield ($mg·L^{-1}·h^{-1}$).
REFERENCES
- [1] Bertalan I, Munder MC, Weiß C, Kopf J, Fischer D, Johanningmeier U. A rapid, modular and marker-free chloroplast expression system for the green alga Chlamydomonas reinhardtii. J Biotechnol. 2015 Feb 10;195:60-6. doi: 10.1016/j.jbiotec.2014.12.017. Epub 2014 Dec 30. PMID: 25554634.
- [2] igem, Registry of Standard Biological Parts, Help:Standards/Assembly/Type IIS
- [3] Gorman, D.S., and R.P. Levine (1965) Proc. Natl. Acad. Sci. USA 54, 1665-1669.
- [4] W.T.Godbey,Chapter13-GeneDelivery,Editor(s):W.T.Godbey,An Introduction to Biotechnology,WoodheadPublishing,2014,Pages275-312.
- [5] Jiang Si, Liu Yingying, Tong shaoming.Effects of four antibiotics on growth and photochemical activities of Chlamydomonas reinhardtii[ J ].Biological processes, 2017,15(02) : 13-20.
- [6] Gao Xiang, Zhou Weicheng, Zhang Fengge, Chen Lanzhou, Wang Gaohong. Effects of four antibiotics on growth and photochemical activities of Chlamydomonas reinhardtii[J]. Plant Physiology, 2015,51(11) : 1887-1894. DOI: 10.13592/J. CNKI. PPJ. 2015.0344.
- [7] Zhu Genmiao, Yang Weiqing. Quorum sensing system regulation on bacterial resistance[J]. Chinese Journal of antibiotics, 2011,36(01) : 7-10. DOI: 10.13461/J. CNKI. CJA. 004713.
- [8] Bertalan I, Munder MC, Weiß C, Kopf J, Fischer D, Johanningmeier U. A rapid, modular and marker-free chloroplast expression system for the green alga Chlamydomonas reinhardtii. J Biotechnol. 2015 Feb 10;195:60-6. doi: 10.1016/j.jbiotec.2014.12.017. Epub 2014 Dec 30. PMID: 25554634.
- [9] Huang Yaping. Construction of nucleus transformation system and expression study of two exogenous proteins in Rhinophyllum (Chlamydomonas reinhardtii) cells. Sichuan University of Science & Engineering,2019. doi:10.27703/d.cnki.gsclg.2019.000015.

